Weeknotes: 27th January 2025
Last week
I was up on The Wirral last week, so got to work from DoES Liverpool for a couple of days, which is a great place to hang out as a maker-space with co-working facilities, and home to many nice people working on interesting things.

Debugging maps
I've still been debugging maps a lot of the last week. Not the same bug, but when you look under a rock you often find other threads to pull on (and metaphors to abuse), and so I'm still down that rabbit hole trying to convince myself that my code is good and my issues are now down to misunderstandings on data. Currently we're at the stage where we have two sources of the same data that are conflicting and I need to work out which is correct.
I did discover that the IUCN redlist data has both global and regional assessments, thanks to help from Simon Tarr at the IUCN. I'd just assumed all species assessments were global, so were confused to find multiple "latest" assessments. The more you know and all that.
Yirgacheffe API improvements new and old
A while ago I wrote a Python geospatial raster processing library called Yirgacheffe, the idea of which is to treat maps (both raster and vector) as things you apply math to directly, rather than working on each pixel. This allows Yirgacheffe to do two things for you:
- Deal with all the geometry offset math when using layers that don't align out the box
- Worry about resource management like how much memory to use and how many CPU cores to use
It is very much a work in progress, and I add things as I need them, and so whilst I could do basic math operators on maps to generate new maps like so:
area_of_habitat = filtered_habitats * filtered_elevation * species_range
Where the first two are raster maps and the last one a polygon map, and the result here is a raster containing only the pixels that are true in the three input layers, like so:
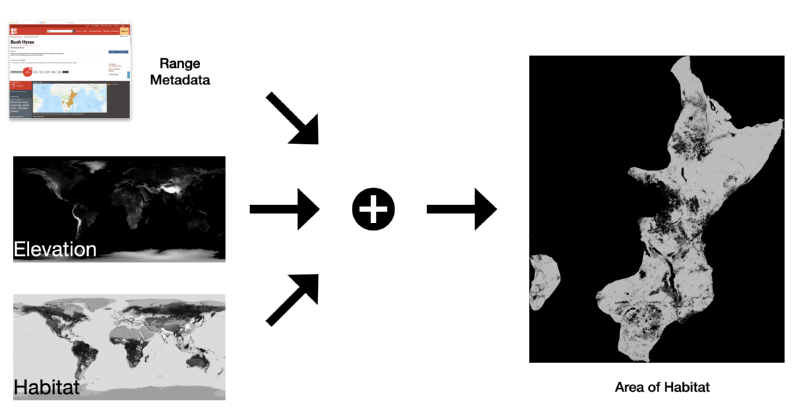
However, some other operations I was doing on maps in my AoH code were much more cumbersome as I'd not added enough logic to Yirgacheffe to do everything I'd needed at the time. When I started yirgacheffe I had however added an escape hatch for myself, allowing me to use Python lambdas that would be passed chunks of data as numpy arrays. So I ended up with code like this in the same file as the above example:
filtered_elevation = min_elevation_map.numpy_apply(lambda chunk: chunk <= elevation_upper) *
max_elevation_map.numpy_apply(lambda chunk: chunk >= elevation_lower)
Over time I have actually added a bunch of new operators to Yirgacheffe as I've used it for more and more things, adding operators such as boolean and/or for example, but never gone back and tidied up the code for earlier projects that uses Yirgacheffe to take advantage of those new features. So I finally caught up examples like the above code, and now it looks like:
filtered_elevation = (min_elevation_map <= elevation_upper) & (max_elevation_map >= elevation_lower)
Here the variables ending in _map are rasters, and the elevation bounds are just integer values, and I end up with a new binary raster that is the elevation between those bounds. The new code is, to me, a lot more clear in what's happening, which is a major point of using Yirgacheffe.
This week, for reasons that I'll go on to in the next section, I decided to go through all the code in my LIFE and STAR implementations (and my AoH library that both use) and find where I was still using my numpy_apply escape hatch, and add whatever operators from numpy I was using to Yirgacheffe itself. The result is that now the code is hopefully more readable and understandable, and it also means that the code should be less buggy, as Yirgacheffe has reasonably good test coverage, so the more work we push into that library the more confidence we can have in it. Removing numpy_apply from the API usage also frees me up to make some additional changes that I've wanted to do for a long time, because it means that I don't have to assume that it's numpy that's doing the heavy lifting on the backend, Yirgacheffe can use other options there...
Metal and CUDA support in Yirgacheffe
When I first started Yirgacheffe I had a version that could optionally use cupy, NVIDIA's Python library for supporting GPU accelerations via CUDA. I had to drop that eventually, in part because I was directly calling numpy a lot of the time.
More recently, because I've been running the LIFE pipeline a lot more, I wanted to both try speed it up on our AMD server that has a NVIDIA GPU, but also on my Apple M3 Max Macbook Pro. I found MLX, which is similar to cupy, in that it has a similar interface to numpy, but uses Apple's Metal GPU framework under the hood. The theory is that, internally Yirgacheffe uses numpy for number crunching, and now that the public API no longer exposes that, I could potentially swap that out for either cupy or MLX when I detect a GPU is present.
As an experiment, I did a hatchet job on a branch on Yirgacheffe and just replaced all the numpy calls with their MLX equivalents and did appropriate remapping of inputs and outputs to numpy (as GDAL still wants numpy data). On my laptop I found for some workloads it made no difference, and for some I got a performance increase of 50% or so. I'm sad that some workloads didn't improve, but they at least didn't get worse. But it's certainly enough to make me want to try get the back-end swapping into Yirgacheffe real soon now. My laptop MacBook Pro isn't ever going to rival the AMD EPYC server I use for most things, as it has 128 CPU cores (256 hyperthreaded) vs 16 CPU cores and 40 GPU cores on my laptop, but given our servers are shared resources, it'd be nice to have some better capability to run these things locally.
Opam
I stated last week that I wanted to start getting some of my OCaml packages into Opam, but I failed to achieve that in the last week, or even get started on that, but I did a note from the original author of the OCaml H3 library which hadn't been updated in many years and I recently got working that he was happy for me to take over maintenance, and so I did some final updates and moved the project to the Geocaml project.
The reason I didn't go further is I know that it needs up to install H3 in order for the package to build, and you can teach Opam about external package dependancies, but I've not had time to dig into how to do that, so for most people the package will just fail to install.
GDAL related admin
In all the playing around with GDAL, I was somewhat surprised that I didn't hit the dreaded error that has plagued me using GDAL from pip since I started using it a few years ago:
...
ImportError: cannot import name '_gdal_array' from 'osgeo' (/home/mwd24/venvs/life/lib/python3.12/
site-packages/osgeo/__init__.py)
I find this problem to be sufficiently common and annoying that I wrote a container system to wrap Python for my ecology colleagues so they didn't need to deal with trying to install GDAL themselves. I suspect a more sensible appraoch would have been to move them from pip to Conda, as Conda seems to do a much better job with GDAL, or give in and use uv like the cool kids do, but that's a whole other podcast.
Anyway, on my Mac I was making and reseting my virtualenv often, and noticed that GDAL 3.10 seems to not hit that issue, where as with the Ubuntu 24.04 LTS I definitely still do. So I've now built GDAL 3.10.1 and plan to upgrade the AMD EPYC server I use to run my puipelines that rather than the standard package. Before I do the swap though I need to check who that might impact so other users don't have any nasty surprises. Even if it doesn't help with pip, there are some nice fixes that it'd be good to have from a newer GDAL release.
This week
Yirgacheffe
Get the CUDA/MLX code to be swappable on the Yirgacheffe backend with the CPU based numpy.
LIFE
Continue trying to get Ali the data she needs for the LIFE follow on paper.
STAR
I said in last week's notes that I wanted to try pickup the work on my STAR metric implementation, but other things got in the way of that.
Talks
On Thursday I'll be one of the speakers at Geomob London.
Tags: weeknotes, life, yirgacheffe, mlx, cupy